Difference between revisions of "Projects:2016s1-120 Attacking Cancer with Signal Processing"
(→Sine Wave Fitting) |
(→Monte-Carlo Analysis) |
||
| Line 87: | Line 87: | ||
The p-value is formed below: | The p-value is formed below: | ||
| − | p(false-positive)=∑(P_REAL<P_VIRUAL)/N | + | p(false-positive)=∑(P_REAL<P_VIRUAL)/N |
Where P_REAL is the peak of real CRP data ,P_VIRTUAL is the peak of Gaussian random pseudo-data and N is the times of simulation. | Where P_REAL is the peak of real CRP data ,P_VIRTUAL is the peak of Gaussian random pseudo-data and N is the times of simulation. | ||
Revision as of 14:28, 26 October 2016
Project Information
Topic: Attacking Cancer with Signal Processing
Supervisors:Dr. Andrew Allison
Adviser: Prof. Derek Abbott
Project members: Jin Hu , Mohammed Said Al-Wahaibi
Introduction
Cancer is one of the most devastating unsolved medical problems. Only 7% of cancer patients on average have a hope of recovery. New approach to solve the problem is to fight the cancer by strengthening the human body's own immune system, by improving the timing of treatment, using signal processing. CRP is produced by the liver and adipocytes in response to inflammation. People who infected by cancer have a different CRP level comparing to healthy ones. Study showed that the best time to apply the immune therapy is when the CRP level is low.
Motivation and Objectives
The main focus of the project is to improve the existing treatment, by improving the timing. The project uses signal processing to estimate the optimal treatment time. The ultimate goal is that this project will help extending human lives.
Previous Studies
In 2009,Dr. Brendon Coventry and his colleagues used Low-Reactive Protein (L-CRP) test to obtain high sensitivity data of CRP. And they found the CRP levels are periodic with the cycle of 7 days.
In 2014, Dr. Mutsa Madondo and his colleagues did their research ,they used Enzyme-Linked ImmunoSorbent Assay(ELISA) to obtain blood samples from patients at seven different times in 12 days' period.Then they claimed that CRP levels and Treg and Teff frequencies did not appear to be oscillatory.
Background
The CRP level of a cancer patients is differing from a healthy human. As the CRP response to inflammations, the change in it level might be periodic and accrue in cycles. The CRP data we have a noisy and irregularly sampled. To separate the noise from the signal we use Fast Fourier Transform techniques and Lomb Periodogram. Both methods are valid way to separate the noise from the signal. We use both method to make sure of getting a valid signal and reducing the possibility of false positives.
Noise Floor
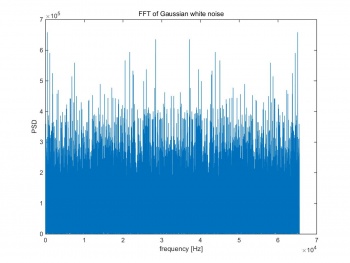
The noise floor is the Fourier transform of noise and unwanted signal. Figure 1 shows the FFT result of Gaussian white noise, which is very noisy and no pure peak can be found. If performing FFT on a noiseless signal , a pure peak will appear on the power spectral density (PSD). For noisy signal, even the peak can be obtained on PSD, the FFT of noise still shows in the background.
Kolmogorov–Smirnov Test
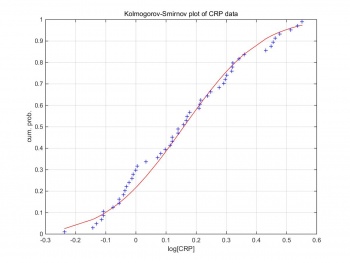
Kolmogorov–Smirnov test can compare a sample with a reference probability distribution. Figure 2 shows the Kolmogorov–Smirnov test of the raw CRP data. The CRP data are formed in log scale,and follow the Gaussian distribution. Therefore, we can generate Gaussian random pseudo-data by creating Gaussian random variables based on the log scale CRP data.
pseudo_data=(random×σ)+μ
De-trending CRP Data
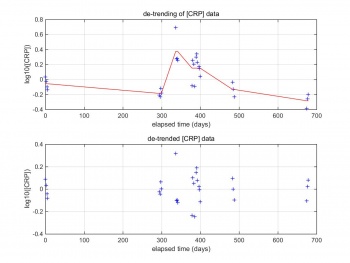
By taking out the trend from our CRP data, it enables us to focus our analysis on the fluctuations in the data. A linear trend typically indicates a systematic increase or decrease in the data, which gives a method for analyzing shorter-term cyclical patterns. These patterns can then be used to more effectively identify major turning points in the longer-term cycle which is what this project is aiming for.as mentioned before the data being treated as separate monitoring period (MP) if the measurement been taken seven days apart. Figure 3 shows the CRP reading for patient No.10. The red line of the top one is the DC component which that go in the middle of each monitoring period. So by taking the DC component will help us identify cyclical patterns. The bottom image is CRP data after removing the DC component.
Analysing the CRP Data
Power Spectral Density
There are two methods to estimate the Power Spectral Density (PSD), one applies fast Fourier transform (FFT) of the re-sampled CRP data, another is using Least-squares spectral analysis (LSSA) of the raw CRP data.
Fast Fourier Transform
The technical challenging of this project is the available CRP data is irregular. When using Matlab to apply FFT of a signal, the samples should be uniform space. So the raw data needs to be interpolated values by spline method, Kriging interpolation or basis function.Spline method can fit the raw data well, but the overshoots is a big issue that may cause the FFT result distorted. In terms of Kriging interpolation, it is too complex to complete the interpolation processing.Even the basis function does not fit the raw data as well as the spline method,the result of basis function is feasible. Moreover, basis function can remove the overshoot which reduces it to the mean level of log scale CRP data. So we Gaussian basis function is the best choose for re-sampling.
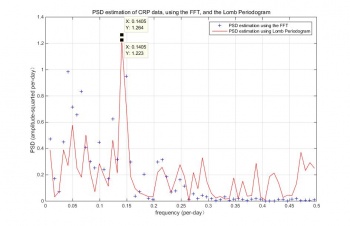
FFT is a useful analytical tool applied in diverse fields, as an effective computational method to calculate a Fourier transform. When analyzing a signal, FFT decreases the number of calculations in order to quickly generate a Fourier transform. So, the properties of FFT and noise processes is well understood. Moreover, FFT and Rayleigh Energy Theorem can use for checking the normalization of calculations. In Figure 4, the blue points are the PSD estimation results of performing FFT. There is an observable peak located at the frequency of 0.1405 per day and the periodic of the peak is 7.117 days.
Lomb Periodogram
LSSA is also called Lomb periodogram, which can apply for irregular samples without re-sampling the data or invent other values . The red line in Figure 4 is the PSD estimation results using Lomb periodogram. The peak locates at the frequency of 0.1405 per day and the periodic of the peak is 7.117 days.
Even though there are some differences between the two results in vision, the peak frequencies of FFT and Lomb periodogram are similar. Both the peak frequencies locate at 0.1405 per day and the periodic at peaks are close to 7 days. So, these two methods are feasible for estimating PSD of log scale CRP data. However, some CRP values are estimated when performing FFT. The data used in Lomb periodogram are all measured value. So,Lomb Periodogram have more details data comparing to FFT as there might some data lost in the re-sampling process before using the FFT.
Sine Wave Fitting
The equation of fitting cure is:
y_i=Acos(ωt )+Bsin(ωt )+C
The coefficient A, coefficient B , constant offset C ,ω and initial parameters are the unknown value will be obtained by using the least square method. The frequency of peak will be used to create the initial parameters. According to the fundamental theorem of algebra, each monitoring periodic should contain at least 5 data for sine-wave fitting.
Based on the periodicity of the sine curve, we can predict the time of the next minimum by using the last fitted data.
The minimum is at
ωt+θ=2Nπ-2π
To estimate the next minimum time,
N= ceil((ωt_Last+θ+π/2)/2π)
The next minimum time:
t_min=(N∙2π-π/2-θ)/ω
Monte-Carlo Analysis
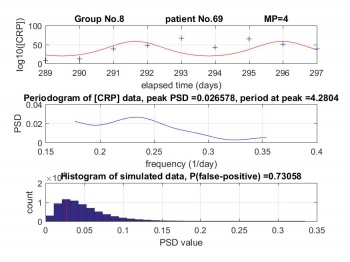
Performing Lomb periodogram of the virtual data to find the peak on PSD. After several times simulation, we can obtain a histogram of virtual data peak distribution.The third image of Figure 5 is the histogram of MP 4 of patient No.69, that simulate for 65,536 times. Enormous times of simulations can obtain a complete histogram. However, for relative long simulation time, there is not significant effect on the histogram.
The probability of false-positive (p-value) is a function of the observed sample results relative to a statistical model, which measures how extreme the observation is. P-value provides information about whether a statistical hypothesis test is significant or not.
The p-value is formed below:
p(false-positive)=∑(P_REAL<P_VIRUAL)/N
Where P_REAL is the peak of real CRP data ,P_VIRTUAL is the peak of Gaussian random pseudo-data and N is the times of simulation.
Results
Testing of Dr. Coventry 's Data
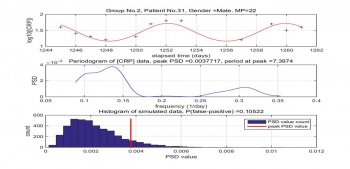
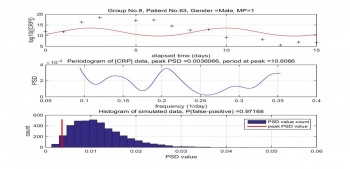
Dr. Coventry found the CRP levels are periodic with the cycle of 7 days. However, after processing the patients' data, we found some periodic at peak is not around 7 days and the peaks are found with the low probability of real. The Figure 6 shows the result of patient No. 31 MP 22. The periodic of peak is colse to 7 days with relatively low probability of false positive.This result is consistent with Dr. Coventry finding. However, the result of Figure 7 is opssite to his finding. The periodic of peak is 10.6 days and with high probability of false positive.
Testing of Dr. Madondo's Data
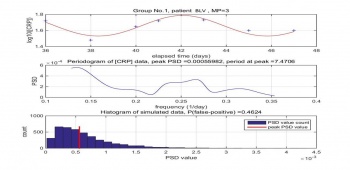
Dr. Madondo's raw data include two groups, IRS and BLV. Performing same signal processing method, we found the data of some MP have high-reliability peak ,which inconsistent to Dr. Madondo's conclusion, the CRP level does not appear to be periodic.