Projects:2018s1-141 CSI Adelaide: Who killed the Somerton Man?
Contents
Supervisors
Honours students
Abstract
The corpse of a mystery man’s body was found at Adelaide’s Somerton Beach, South Australia, Australia, on the 1st December 1948 and was hence referred to as the Somerton Man. Till this day the identity of the Somerton Man and the cause of his death is unknown. This project will be broken up into three different tasks, with all contributing towards the unsolved case of the mysterious man. The first task is based around the piece of paper that was found inside his trousers pocket. This piece of paper had five lines of capital letters, and is thought to be some kind of code or cipher. It was found that this mysterious code was a part of a poetry book, known as the ‘Rubaiyat of Omar Khayyam’. The letters on the mysterious code are thought to be first letters of words, based on previous year’s project groups. The location of his death is near Morphettville Racecourse, which leads to belief that the Somerton Man’s mysterious code are different horse names. However, using various statistical approaches, this was proven to be not true. The second part of this project involved, using a mass spectrometer to analyse different isotopic signatures of the samples. More specifically, the samples used were the shaft of the hair, obtained from various people, which were ablated by the laser, and effectively recorded the various elements. This will be compared with the Somerton Man’s hair, to identify specific elements present, as well as noting how long he was in Adelaide before his death. Different DNA samples, were analysed in the final task. Using software tools the samples were degraded, until the DNA became unidentifiable. It was found that degrading 50% or greater of the DNA sequences, makes the DNA unidentifiable.
Acknowledgements
Project supervisor Professor Derek Abbott, for the helpful and motivational advice, as well as the exceptional guidance, which was presented on each of the completed tasks. The University of Adelaide Microscopy Centre, for permitting the project team to use the mass spectrometer. Dr Sarah Gilbert for the assistance and suggestions whilst using the mass spectrometer. Also, thank you to all the volunteer who gave their hairs to use during the experiment. In addition, the project team would like to acknowledge all the software support from University of Adelaide, including Microsoft, Matlab as well as Iolite from the Microscopy centre.
Introduction
Motivation
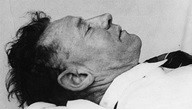
During this project, a study was undertaken on an unsolved murder case. On December 1st, 1948, an unknown man was found deceased, on Somerton Beach in Adelaide [1]. From then on, he was labelled, ‘the Somerton Man’. There was no form of identification present on him, as well as little information surrounding his death. Therefore, the cause of his death is still unknown to this day [2]. Figure 1 shows the deceased man.
A piece of paper with the words “Tamám Shud” printed on it, was found rolled up inside his trousers pocket, which can be seen in Figure 2. This statement can be translated from Persian to either “it is ended” or “it is finished”. This piece of paper was found to be part of a poetry book, the ‘Rubaiyat of Omar Khayyam’ [3]. The theme of the book is that, one should live life to the full and have no regrets when it ends [4].
The book is speculated to be related to the dead man, due to the parchment. Hence, the case being known as the Tamam Shud case. This has been considered, since the early stages of the police investigation, to be "one of Australia's most profound mysteries [6].” Capital letters were found to be scribbled in the back cover of the Rubaiyat of Omar Khayyam, as seen in Figure 3. Thus, indicating that these letters are somewhat vital to the case at hand, as it is speculated that they may be a form of code or cipher.
The code consisted of five various lines of capital letters, with a total of fifty letters all together. The second and fourth lines are very similar in the way in which they appear. It is therefore, believed that the Somerton Man may have made an error with the encryption, when writing the second line, hence why it is struck out. It is unclear whether some of the letters are in fact an “M” or “W” [7]. For tasks referring to the code, it is assumed that the unclear letter is an “M”. His body was found to be located near the Morphettville Racecourse, this leads to the belief, that the Somerton Man’s mysterious code are related to different horse names. It was also noted that the people who found the deceased body of the Somerton Man, were racehorse jockeys [2]. An Inductively Coupled Plasma Mass Spectrometer (ICP-MS) is used in this project to identify the various isotopes in various people’s hair. With the main concern of Strontium as, Adelaide has high levels in strontium in the soil compared to the rest of Australia.
Previous Studies/Related Work
Australian Department of Defence
In 1978, a request was sent by journalist Stuart Littlemore to the Department of Defence cryptographers to analyse the code. Unfortunately, the cryptographers were unable to crack the code, as they could not provide a satisfactory answer. It was stated that the code had “insufficient symbols” or a “disturbed mind” generated the meaningless code [8].
The University of Adelaide project groups
There have been several Honours project groups at the University of Adelaide that have undertaken this project. The previous work the project groups have done include:
- Letter frequency analysis in different languages.
- Initial letter and sentence letter probabilities.
- The likelihood of the code being an initialism of a poem.
- Different cypher techniques.
- The design and implementation of a web crawler.
- 3D generated reconstruction bust of the Somerton Man.
Main conclusions that these project groups have come to are:
- It is unlikely that the mysterious code is created randomly.
- There is strong evidence to believe the mysterious code is most likely to be in English.
- It is not likely that the mysterious code are initialisms extracted from poems.
- The Rubaiyat of Omar Khayyam was not used as a straight substitution one-time pad for encryption.
- The Rubaiyat of Omar Khayyam was not created as a one-time pad for the mysterious code.
With these conclusions, this project will look into further detail of what the Mysterious Code is [9] [10] [11] [12] [13] [14] [15].
Mass Spectrometer Pervious Work
Previous years have also done study with Mass Spectrometer. The 2013 project group had some of Somerton Mans hair and plotted the different elements in the hair comparing with controlled samples. Analysis was conducted on different elements between the two samples. This was done using a glass slide, which have impurities in it [13]. In the 2016 project group, they recreated the analysis using a quartz slide, which does not have impurities. They concluded that Somerton Mans had some abnormally high readings, of some elements, one of which is strontium [15]. In this project, the strontium level will be looked in higher detail and this will be used to indicate how long the Somerton Man was in Adelaide before his death.
Aims and Objectives
The first task that will be done is to understand if the mysterious code are a collective object (horse names, Adelaide street names, Australian beaches, etc.). This will be done using hypothesis testing. An extension of this task will also be completed, which involves the Rubaiyat of Omar Khayyam. The second task that will be performed is using a mass spectrometer. Controlled sample hairs will be compared with Somerton man’s hair, to see how long he was in Adelaide before his death, by finding different elements in the hair. The third task that will be accomplished is using DNA data. The data will be degraded using software tools till it becomes unidentifiable. This then can be used to see how much DNA we need from Somerton man, where further research can undergo.
Technical Background
P-value Theorem
A p-value is a recognised statistical probability, which acknowledges whether an equal or larger effect is present, in comparison to its observed counterpart. In statistics, the p-value helps you determine the significance of the statistical hypothesis by observing the results that were sampled. This determines the probability that the results are due to chance, rather than the experimental conditions. Thus, determining the strength and validity of the results against that of the null hypothesis [16]. In this project, the main focus with the p-values is to determine whether the mysterious code, represents local horse names. Where the null hypothesis is ‘The group of letters are horse names’ with the alternative hypothesis being ‘The group of letters are not horse names.’ For null hypothesis to be accepted the p-value must be larger than 0.05, this indicates that the observed data point is located in the ‘most likely observation’ range, as seen in Figure 4. If the p-value is lower than 0.05, this indicates that the collected results are statistically significant and that the observed data point is located in the ‘very unlikely observations’ range. If the results are in the ‘very unlikely observations’ range, then the null hypothesis can be rejected, which means that the mysterious code is indeed, not horse names [17].
Figure 4 shows, the y-axis is the probability and the x-axis is the set of possible results.
Mass Spectrometer
A mass spectrometer is an analytical technique which, when given a foreign sample, it can detect unknown compounds within it. The mass spectrometer produces a multitude of ions in the sample, which then uses a mass to charge ratio of the different ions and records the quantity of each ion type [19]. The components of a typical mass spectrometer is seen in Figure 5. The three major components are the ion source, analyser and the ion detector system.
The ion source produces gaseous ions from the sample that was used. The analyser, then sorts the different ions by using the mass-to-charge ratio, according to their mass components. The ion detector system detects the different ions in the sample and records the quantity of each ion type and converts it into an electric signal [20]. The Inductively Coupled Plasma Mass Spectrometer (ICP-MS) is the type of mass spectrometer that was used in this project. The ICP-MS is faster, more precise and sensitive at finding different ions compared to other types of mass spectrometer [21]. In regards to the project, the sample being used is the shaft of the hair.
Single Nucleotide Polymorphism (SNP)
Single Nucleotide Polymorphisms (SNPs), are one of the most common and well talked about genetic variations, which can be present between humans [22]. These variations occur within a nucleotide (a single block of DNA) and happen roughly within 1 out of 300 base pairings [23]. In regards to the project, SNPs will be removed from a DNA sample.
Knowledge Gaps and Technical Challenges
The technical challenges that will be encountered in this project are related to all the knowledge gaps mentioned. To complete each task within the project, further development for programming skills, such as Matlab were required. P-value calculation and hypothesis testing needed some revision, to ensure that a satisfactory level of understanding of the concepts was present. The skill to use Microsoft Excel to perform statistical analysis on the p-value, is required. It was also required to learn how to correctly use a mass spectrometer and interpret the results, this can be done by finding multiple ways to enhance knowledge, before trials.
Task 1: Code Analysis
Aim
The aim of this task is to comprehend whether the mysterious code represents some collective object. The collective objects that will be used are horse names, Australian beaches and cities, South Australian street names, and The Rubaiyat of Omar Khayyam book. The assumption will be made that the letters in the mysterious code, are the initialism of a word. The Somerton Man had a lot of associations with horses, so further research will be going into the assumption that the mysterious code are ‘horse names’. The null hypothesis is ‘The group of letters are horse names’ and the alternative hypothesis is ‘The group of letters are not horse name.’
Preperation
Before the initialisation of this task, the details of the whole case were reviewed. This review included the following; who found the corpse, where it was located and when it was discovered. This gave a better understanding of what type of collective object should be researched, which are stated above. As mentioned before, the reason why horse names are a major factor is because the location of his body was found to be located near Morphettville Race course, as seen in Figure 6.
It should also be noted that the people who found the deceased body were racehorse jockeys. Hence the reason we assumed the mysterious code are the initialism of horse names from the year 1948.
Method
The approach to determine if the mysterious code represents collective objects, will be done by calculating the p-value and implementing hypothesis testing. In the case of the horse names, there were no direct websites, which provided horse names in the year of 1948. This led to discovering evidence of these names within relevant newspapers and articles. This was done by using ‘Trove’, a search engine to help find resources in Australia. More specifically, in this case it was used to obtain articles and newspapers from 1948. The other collective objects, as mentioned above, were found using South Australian government websites. This led to an abundance of cross checking, to make sure that the list which was going to be utilised, was indeed correct. Matlab was used as the software tool. The initial letter of the each collective object was all that was needed, where using code was implemented to perform this task. In the case, where a collective object had multiple words, both words were included. Also if there was any extra punctuation, it was removed. A demonstration is shown in Figure 7, which illustrates the input and output results for the horse names case and also displays the frequency of each letter.
Excel was used, to produce the statistical results from Matlab. The letter frequency will be obtained by dividing the amount of each letter over the total amount calculated, where then a p-value test was performed and a comparison graph was completed.
Results
Each collective object was compared to the mysterious code by the frequency of each letter. Where the x-axis represents the alphabet and the y-axis represents the frequency of the letters between the two testing objects. The p-value test was also completed to verify the results, where a p-value of less than 0.05 shows that it is very unlikely that the collective object is the mysterious code.
Horse Names
The comparison of horse names to the mysterious code is seen in Figure 8.
There was a sample of 69 horse names and it can be seen on the graph that the horse names do not correlate with the mysterious code with many of the English letters. This was also proven by the p-value, as it was lower than 0.05, which means the null hypothesis is not accepted.
Australian Beaches
The comparison of Australian beaches to the mysterious code is seen in Figure 9.
There was a sample of 114 beach names. Analysing the graph it be seen that the frequency of the letters do correlate with mysterious code. As the results seemed genuine a hypothesis test was done between this values. The results showed a p-value of greater than 0.05, which indicates that the mysterious code could be Australian beach names.
South Australia Street Names
The comparison of South Australian street names to the mysterious code is seen in Figure 10.
There was a sample of 447 South Australian street names. Observing the graph it can be seen that the frequency of the letter are not similar with the mysterious code. This was also proven by the p-value, as it was lower than 0.05.
Australian City's
The comparison of Australian city names to the mysterious code is seen in Figure 11.
There was a sample of 90 Australian city names. Observing the graph it can be seen that the frequency of some letter are similar with the mysterious code. A hypothesis test was then done to check the results. The p-value that was obtained was less than 0.05.
The Rubaiyat of Omar Khayyam book
The comparison of the Rubaiyat of Omar Khayyam book to the mysterious code is seen in Figure 12.
There was a sample of 852 words form the book. Observing the graph it can be seen that the frequency of the letter are not similar with the mysterious code. This was also proven by the p-value, as it was lower than 0.05. An extension of this task was also done. This includes analysing The Rubaiyat of Omar Khayyam book more carefully. Previous years stated that the mysterious code does not correlate with the book. Each paragraph in the book has four lines of words (see Figure 13), which compared with the mysterious code also has four lines. Still assuming that each letter in the mysterious code is an initial word, we can compare the two.
The task was to count how many words are in each line of the book and compare it with the mysterious code. Using the first paragraph in Figure 13 (outlined with a red square), the first line has 9 words, then followed by 7 words in the second line, then 8 words in the last two lines. Comparing just the first paragraph with the mysterious code from line 1 to 4, there are 9, 11, 11 and 13 letters respectively. It already can be seen from the first paragraph that there may not be a correlation between the mysterious code and the book. Counting every line would be very time consuming, therefore a text file of The Rubaiyat of Omar Khayyam was used, in correlation with Matlab to count each word in each line. Then using excel, a graph was plotted with error bars to the number of letters in the mysterious code. This can be seen in Figure 14.
The x-axis represents which line in the paragraph it is and the y-axis represents the amount of words present in that line. It can be seen that on line 1, the mysterious code is in the error bars. The rest of the lines are out of the error bars. This indicates that the mysterious code is not from The Rubaiyat of Omar Khayyam book and further proves the previous year’s studies of the book not being part of the mysterious code.
P-value
A summary of the p-values are shown in Table 1.
It can be observed that the only collective object that is above 0.05 is Australian beaches, this indicates that the mysterious code could be Australian beaches.
Conclusion
Overall, the results have shown, that it is unlikely that the mysterious code represents the hypothesis stated, of that it is horse names. It has also shown that it is unlikely to be South Australian street names, Australian city names or The Rubaiyat of Omar Khayyam book, even with extensive analysis on the book. But to some surprise it is possible that the mysterious code is Australian beach names, as the p-value was above 0.05. Future research will be required to obtain more meaningful results.
Task 2 Hair Analysis
Background Theory
Analysing elements in the hair could reveal a great deal of useful information on a person’s recent life, this includes their lifestyle imbalances, living environment and dietary problems. Also the mineral levels in hair is about ten times more robust, than compared to blood [26]. These results could reveal where the last place Somerton Man had been to or even the last activity that Somerton Man had done, which could provide some useful evidences in solving this case. Knowing how much hair grows is very important for this project. For every month that passes, hair grows by 1cm [27]. This means the newest hair is in the root.
Aim
The aim of this task is to identify the different isotopes present in several different people’s hair. More specifically the element of concern is strontium. Adelaide has high levels in strontium in the soil compared to the rest of Australia. With this knowledge, the task is to test various hair samples, which have left Adelaide, within the past month and compare it to that of hair samples that have not left Adelaide for at least a year, to see how the strontium values change. This will then be compared with the strontium levels in the Somerton Man’s hair, which can determine how long he was in Adelaide before his death. The ICP-MS, is the approach to determine the different isotopes within the hair, which will then return a spectral analysis of the hair. The spectral analysis will be completed by laser ablation of the hair, where the hair will be ablated with a laser and the spectral elements are recorded.
Preparation
Hair Elements
Before the ICP-MS could be used for the analysis on the hair samples, different isotopes had to be chosen for the ICP-MS to find. Research was completed to find the most common chemical elements inside human hair, which included carbon, hydrogen, oxygen, sulphur, phosphorus and zinc. Then further research to indicate what element we want the ICP-MS to find, which included any toxic elements and common elements in food. There were 24 isotopes that were recorded by the ICP-MS and are shown in Table 2.
Hair Samples
In this project, a different hair samples was obtained, from five different individuals, for more reliable and accurate results. Two of these samples obtained, were from people that left Adelaide for a period of time and the other three samples were from people that stayed in Adelaide. All the samples will be anonymous for privacy reasons and will be identified as A, B, C, D and E. A summary of each hair sample is shown in Table 3, this includes which sample is identified as, sex, the date the hair was obtained and a brief description of the sample.
The length of time out of Adelaide is very important, as we can determine the length of hair that was in Adelaide and the length of the hair that was outside of Adelaide. Sample A left Adelaide for 7 days, assuming there is 30 days in one month and hair grows 1cm per month, therefore 2.33mm of hair is from Japan. Similarly with sample E, they left Adelaide for 13 days, therefore 4.33mm of hair is from Bali. Also have to take consideration of the 2 days from when they came back to Adelaide to when the hair was obtained, this is 0.67mm. An example of the length of the hairs is shown in Figure 15.
The red displays the 2 days before obtaining the hair, when both samples came back to Adelaide. The orange shows the length of sample A and the green shows the length of sample E, when both samples were away from Adelaide.
Quartz Slide
The hair samples will be placed on a slide, in this case the slide will be made of pure quartz. The reason a pure quartz slide is used, rather than an ordinary glass slide is that, glass slides have a lot of impurities that would contaminate the result, where as a pure quartz slide does not. The elements in a glass slide and quartz slide is shown in Table 4.
The hair samples were stuck down with double sided sticky tape on the quartz slide, this will have some contamination and will have to be dealt with appropriately.
Mass Spectrometer Experiment
Capturing the data
The quartz slide was placed into two stabilisers, which was then installed into the machine. This machine did the laser ablation on the hair samples. Figure 16 displays the quartz slide with the hair samples, installed into the laser ablation system. While Figure 17 displays the enlarged version of the quartz slide on the monitor.
Next to the laser ablation system, there are two monitors which will be used for the laser ablation task. The left monitor records the data of the laser ablation and the right monitor controls the laser ablation system, this can be seen in Figure 18. The next step involved choosing how many spots of the hair we want ablated and the distance between these spots. A spot is where the laser will ablate the hair and the isotopes will be recorded for that spot. It was chosen to ablate the hair about half way (1cm), which gave 11 spots at about 800 micro meters apart, two of these spots were located in the root. The mass spectrometer will then document the chosen isotopes. The complete set up of the experiment is shown in Figure 19, with the laser ablation system on the left and the mass spectrometer on the right.
Processing the Data
The data from the ICP-MS was then put into a software called Iolite, this is how we managed the data and removed any anomalies [29]. The overall waveform of the isotopes in the hair sample for several spots can be seen in Figure 20. An enhanced and zoomed in version of one of the spots is shown in Figure 21.
A brief explanation of the waveform composition will be done. Each waveform is an individual isotope, where the top waveform is the average of all the other waveforms. The first 20 seconds of the waveform, shows a relative flat line, this is the laser ablation system calibrating itself. This information is not useful. The waveform peaks up (represented with a square box), this is when the laser just hits the surface of the hair. Then the waveform is a flat line, this is when the laser is ablating the inside of the hair. This is the important information. In some cases the waveform peaks up at the end, this is the laser ablating through the hair and onto the sticky tape and quartz slide. To compare the results more precisely, two sets of data were captured, the surface of the hair and inside the hair. Only getting these results, meant there was so contamination from the sticky tape or the calibration set up. Lastly the results were outputted into one comprehensive Excel file, with all the necessary data, a portion of the data can be seen in Figure 22.
Data Analysis
Analysing the data, there were several isotopes that were found in the hair, these isotopes are shown in Table 5.
Strontium
The comparison of the strontium in the different samples can be seen in Figure 23.
The x-axis represents the distance of the hair ablated and the y-axis is the amount of strontium in the hair at that distance. Samples A and E, are the samples that left Adelaide and came back. For sample E about the first 4000um of hair is when they were in Bali, except for the very first point where they were in Adelaide. Observing the figure, sample E has two high peaks of strontium levels, one at about 1500um and the other at about 3200um, this indicates that there is high strontium levels in Bali. The rest of the value of strontium are low, when sample E came back to Adelaide. For sample A about the first 2000um is when they were in Japan, except for the very first point where they were in Adelaide. Observing the figure, there is a peak at about 1500um, indicating higher strontium values, than that of when sample A was in Adelaide. Overall, these results are concluding that the strontium levels in Adelaide are smaller than that of Bali and Japan.
Conclusion
The strontium level is lower in Adelaide than that of Bali and Japan. Relating this to the Somerton Man, as pervious groups did the ICP-MS on his hair and got high strontium reading. This indicates that Somerton Man must have visited another country with high strontium level, before his visited Adelaide.
Task 3 DNA Analysis
Background Theory
Deoxyribonucleic acid (DNA), is the hereditary material in humans and all other organisms, it is mostly located in the cell nucleus but a small portion can be found in the mitochondria [30]. DNA is made up of four chemical bases, which are adenine (A), guanine (G), cytosine (C), and thymine (T), the information of DNA is stored as a code [30]. More than 99% of these bases are the same between individuals, the variant of DNA determines the different personal characteristics an individual has [30]. Furthermore, this DNA variation is called single-nucleotide polymorphism (SNP), this is an important aspect in this task.
Aim
The aim of this task is to discover how much DNA can be degraded before it becomes unidentifiable. This information will also be used to indicate how much DNA is needed from Somerton Man. The approach to degrade DNA, will be done by removing SNPs from a DNA sample.
Preparation
Obtaining DNA data
A DNA sample kit was ordered from an ancestry place called 23andMe. This is a privately held personal genomics and biotechnology company, which they use saliva to test your DNA [31]. The DNA sample was tested and the results were received. This can be seen in Figure 24.
Figure 24 show details of the DNA report. It can be seen that this person is 65.1% Eastern Europe, indicating at least one of their parents are European. Another useful information is the Maternal Haplogroup and the Paternal Haplogroup. A haplogroup is a group of similar haplotypes that share a common ancestor but different SNPs [32]. The Maternal Haplogroup traces back to a single common ancestor, this can be used for both males and females, which in this case is H11. The Paternal Haplogroup use Y-chromosomes to define a specific set of shared mutation, this can only be used for males, as females do not have Y-chromosomes and can be seen is R-CTS11962.1.
Raw Data
23andMe also provided the raw data of the DNA sample, this document was the basis of this task. Figure 25 shows a portion of the raw data.
Each line in the raw data represents one single SNP. Removal of these SNPs will be done at random. The raw data has over 600,000 lines.
DNA database
After analysing and understanding the raw data, it was then imported into a DNA database website called ‘GEDmatch Genesis’ to compare with other DNA samples. A fraction of the original sample and how the database displays the results is show in Figure 26.
For privacy reasons the names and email address could not be display in the figure above. Each column in Figure 26 will be explained. ‘Kit’ represents different DNA samples from various individuals. A segment refers to a section or block of contiguous SNPs. A matching segment is a section that is the same between two people. ‘Largest Seg’ is the largest matching segment. ‘cM’ (Centimorgan) is a measure of genetic linkage, it is a measure of DNA information within a chromosome. ‘Gen’ provides a rough estimate of the number of generations between you match both share, where 3 indicates two candidates share the same great-grand parents, 4 indicates they share the same great great-grand parents etc. 'Overlap' is the number of positions that exist in common between both kits, without regard to whether they match or not. The amount of overlap, along with the largest cM amount, is usually a good indication of the relative quality of the match. Matches with low overlap are highlighted with a pink or red background, depending on the overlap value. ‘Date compared’ the date the DNA sample was processed into the database. ‘Testing company’ the location where the DNA sample was tested.
DNA Degradation
Degradation in this case refers to the randomly removal of DNA sequences form the raw data. With these results, SNP’s will be removed from the DNA at different percentages, which then can be utilised to discover how much DNA is required until it becomes unidentifiable. Matlab was used to remove different percentages of SNPs in the sample. Figure 27 demonstrates the code that was used to remove the SNPs from the raw data.
A brief explanation of how the code works will be discussed. The code gets the raw data (which is a text file). A different portion of the raw data is shown in Figure 28, which will be used in a demonstration.
The raw data has 638468 lines, the code then removes a certain percentage of those line (The code is currently going to remove 90% of the lines). The next part of the code deletes the blank space of the removed lines. The blank spaces needed to be removed to import the data to GED match Genesis. A demonstration of how the code works is shown in Figure 29.
10 lines are present in the raw data (Left image), then 2 lines are removed (Middle image), lastly the blank spaces of the removed lines are also removed, leaving only 8 lines in total (Right Image). Lastly the code converts the results back into a text file, which can be imported to GED match Genesis. The removal of SNPs was done from 10%-90% and the whole set of the experiment will be carried out five times to obtain an average result to reduce any outliers. Two different task were completed using the DNA, more specifically the degraded DNA. One tasked involved using heritage analysis and the other involved comparison of DNA using the database.
Heritage
The first task completed was investigating the heritage of the DNA sample sent. There were various genetic ancestry projects that could have been chosen, therefore research was completed on all the different genetic ancestry projects to make sure that the results would be the most accurate. Eurogenes seemed the most appropriate considering the sample is decent of European background. The next part was to select which model was most appropriate to calculate the heritage in the DNA, again significant research was completed and the chosen model was Eurogenes K13, as this model is best for samples with mixed heritage. The heritage results for the original case (no SNPs removed) can be seen in Figure 30, it can be seen that the sample has strong heritage in North Atlantic, Baltic and West Mediterranean regions.
The results that were completed only took consideration of North Atlantic and Baltic regions as these had the highest percentages, 29.13% and 42.11% respectively. The other cases were than completed, the 10%-90% of SNP removal. A graph was produced to visualise what was happening when removing the SNPs, which can be seen in Figure 31.
The x-axis represents how much SNP is removed in the sample and the y-axis is how much percentage of that heritage the sample is. Observing the graph it can see the results are relative steady up to about 60%, the results then fluctuate at 70% and onwards. This indicates that removing more than 70% of the SNPS will result in inaccurate results. This result is not enough to conclude this, so five more tests were completed to see if the trend was similar for each case. The figure below demonstrates the average of the heritage tests completed.
It can be seen that average results are all relatively linear, expect for some points at North Atlantic 40%, 80% and 90% and Baltic 80% and 90%. The error bars however, get larger as the percentage of SNPs are being removed. This indicates that the DNA samples start to lose its structure as SNPs are being removed. From 10%-40% of SNP removed, the standard deviation starts increasing to approximately 1, which shows the DNA is still robust. From 50%-90% of SNP removed, the results starts to vary significantly, which shows as the error bars are large and the standard deviation is much larger. It can be concluded with these results that DNA sample is robust until about 50% and after that DNA becomes unidentifiable.
Database
The second task that was completed was comparing the DNA sample to a database of other people DNA samples. Using the database, we were able to efficiently identify how closely related, the DNA samples were, to other people. Refer back to Figure 26 to see how the database displays the results. The original sample will be compared with the 10%-90% cases of SNP are removal. The objective is to find false positives and false negatives. A false positive in this case would mean that the kit appeared in the removal section and a false negative means the kits disappeared in the original sample. An example below will be shown for a clear understanding. Original: A B C D E 10% of SNP removed: C D G H I false positive: G H I and false negative: A B E As there is thousands and thousands of kits to compare, a sample size of 30 was taken to compare. Figure 33 shows the false positives and false negatives for different percentages of SNPs removed.
The x-axis represents how much SNP is removed in the sample and the y-axis is how many false positives or false negatives are found. Unfortunately, in this case the false positives and false negatives equalled each other. The results are all fairly high, at 10% there were only 6 matches with the original case. This indicates that even altering the DNA just by a small amount can have significant change on the DNA. It can be seen as it starts to approach 50% of SNP removal there are more false positives and false negatives, with fewer matches to the original case. At 50% and greater of SNP removed there is 30 false positives and false negatives, this means that no other DNA sample matched the original case.
Conclusion
The major finding in this task is that when 50% of the DNA is removed it becomes unrecognisable. This was proven in both the heritage task and the database task. It was also found that alerting the DNA by only a small amount can result in very significant change to the DNA. Relating this back to the Somerton Man, only 50% of DNA is needed from the Somerton Man to undergo further research.
Project Management
Budget
Each project member had $250 for the project, hence we had $500 for the entire project as two members were present in the group. The pure quartz slide was purchased for the mass spectrometer The DNA genetic testing kit was purchased so we could test our DNA. A table of what items were purchased and how much each item cost is seen in Table 6.
Risk Management
A risk assessment table for the project can be seen in Table 7. One of the major risks that was encountered was bugs in the code. This caused lots of issues throughout the project, one of the main issues was that it delayed the project from proceeding further, this lead to task completion delay. Another risk was the chance of having the Human Ethics not approved. This would mean that we could not do the mass spectrometer task and the DNA task, meaning we would have to do more in-depth analysis on the first task and find additional tasks to do. Slight misunderstanding of project tasks happened throughout the project, but these were explained through meetings with the supervisor.
Conclusion
The main task was to find out if horse names represent the mysterious code. Final conclusions for the code analysis task, showed that the mysterious code is unlikely to be horse names. It was also discovered that is unlikely to be South Australian street names, Australian city names or The Rubaiyat of Omar Khayyam book. Even with further studies on the book, it was still unlikely to be the mysterious code. But the results showed that it is possible that the mysterious code is Australian beach names. The second main task was ICP-MS on different hair samples. The main focus was strontium and was compared with two samples that left Adelaide for a period of time and the other three samples that stayed in Adelaide. It was found the level of strontium was higher in Bali and Japan than Adelaide. This concludes that Somerton Man must have visited another country with high strontium before coming to Adelaide. The last task involved degrading DNA to discover when the DNA would become unidentifiable. This was done by removing SNPs. Main conclusion found was that DNA become unidentifiable, when 50% of the DNA is removed. This could be used to help find the Somerton Mans relatives or his real identification. Overall, the possibility of discovering the identification of the Somerton Man has gotten a step closer. Hopefully with this project completed other groups can utilise this information and identify who the Somerton Man is.
Future Project Discussion
Code Analysis
Deciphering the mysterious code still needs to be completed. It is possible that the mysterious code is Australian beach names, therefore further investigation is required to finalise if this is true or not. This can be done by increasing the sampling size or by locating what beaches Somerton Man might have visited before the time of his death. Also, more collective objects should be considered and researched. Using different statistically methods would also enhance the results.
Hair Analysis
Analysis of hair from different countries that the Somerton Man could have gone to and compare those results to the strontium levels of this project. To confirm that the hair sample didn’t have any outliers in it, each individual should give two hairs to sample and the whole hair should be ablated rather than doing spots in the hair. This will be more accurate, as you will have more data to process.
DNA Analysis
With the DNA task, as there was only a small sample of false positives and false negatives used, an algorithm or code should be used to process all the false positives and false negatives from the database. This would give a more accurate result. Another task would be having a second DNA kit from another individual and compare the two together. This will see if the two DNA results, have the same undefinable DNA percentage. Through extensive research on the case, Somerton Man has a granddaughter. Future groups can research on how the DNA sequence is different between a grandfather to their granddaughter. This could help understand the different DNA sequences between the two. Now that we know that only 50% of DNA is needed, future groups should get Somerton Mans DNA and undergo further research on it.
References
[1] J. Bineth. “Somerton Man cold case,” ABC news, 14th December 2017 [online]. Available: http://www.abc.net.au/news/2017-12-14/somerton-man-cold-case-could-be-one-step-closer-to-solved/9245512
[2] The News, “Dead Man Found Lying on Somerton Beach,” 1st December 1948, p. 1 [online]. Available: http://trove.nla.gov.au/ndp/del/article/129897161
[3] The Advertiser, “Cryptic note on body,” 9th June 1949, p. 1 [online]. Available: https://trove.nla.gov.au/newspaper/article/36371152
[4] The Advertiser, “New Clue in Somerton Body Mystery,” 25th July 1949, p. 3 [online]. Available: https://trove.nla.gov.au/newspaper/article/36677719
[5] “Tamam Shud/ Somerton Man,” [online]. Available: http://ciphermysteries.com/tamam-shud-somerton-man
[6] The Advertiser, “Tamam Shud,” 10th June 1949, p. 2 [online]. Available: https://trove.nla.gov.au/newspaper/article/36371416
[7] Maguire, S. “Death riddle of a man with no name,” The Advertiser, 9th March 2005, p. 28 [online]. Available: http://www.eleceng.adelaide.edu.au/personal/dabbott/tamanshud/advertiser-mar2005.pdf
[8] Inside Story, presented by Stuart Littlemore, ABC TV, screened at 8 pm, Thursday, 24th August, 1978
[9] A. Turnbull and D. Bihari. (2009). Final Report 2009: Who killed the Somerton man? [online]. Available: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Final_report_2009:_Who_killed_the_Somerton_man
[10] K. Ramirez and L-V. Michael. (2010). Final Report 2010 [online]. Available: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Final_Report_2010
[11] S. Maxwell and P. Johnson. (2011). Final Report 2011 [online]. Available: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Final_Report_2011
[12] A. Duffy and T. Stratfold. (2012). Final Report 2012 [online]. Available: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Final_Report_2012
[13] L. Griffith and P. Varsos. (2013). Semester B Final Report 2013 – Cipher Cracking [online]. Available: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Semester_B_Final_Report_2013_-_Cipher_cracking
[14] N. Gencarelli and J. Yang. (2015). Semester B Final Report 2015 – Cipher Cracking [online]. Available: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Final_Report/Thesis_2015
[15] Y. Li and Y. Ma. (2016). Semester B Final Report 2016 – Cipher Cracking [online]. Available: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Final_Report/Thesis_2016
[16] B. David. “P Value and the Theory of Hypothesis Testing: An Explanation for New Researchers,” Clinical Orthopaedics and Related Research®, Vol.468 (3), pp.885-892 2010.
[17] G G. L et al. “What is the Value of a p Value?,” The Annals of Thoracic Surgery, Vol.87(5), pp.1337-1343 2009.
[18] T. Hung. “The p-value,” 21st March 2016 [online] Available: https://www.students4bestevidence.net/p-value-in-plain-english-2/
[19] “Mass Spectrometer,” Premier Biosoft [online]. Available: http://www.premierbiosoft.com/tech_notes/mass-spectrometry.html
[20] R. Bakhtiar. “Biological mass spectrometry: A primer”. Mutagenesis pp.530-531 2000.
[21] S. Jackson et al. “The application of laser ablation-inductively coupled plasma-mass spectrometry to in situ U–Pb zircon geochronology,” Chemical Geology, Vol. 211, pp.47-69 2004.
[22] “What are single nucleotide polymorphisms?,” U.S National Library of Medicine [online] Available: https://ghr.nlm.nih.gov/primer/genomicresearch/snp
[23] G. Shaw. “Polymorphism and Single nucleotide polymorphisms (SNPs)” Science Made Simple, Vol. 112, pp.664-665 2013.
[24] Google Maps, 2018. [online]. Available: https://www.google.com.au/maps/dir//Morphettville+Racecourse
[25] E. FitzGearld. “Rubaiyat of Omar Khayyam,” [online]. Available: http://www.eleceng.adelaide.edu.au/personal/dabbott/tamanshud/W&T_rubaiyat_wells_copy.pdf
[26] Dr. W. Lawrence. “HAIR MINERAL ANALYSIS – AN INTRODUCTION,” [online]. Available: https://www.drlwilson.com/articles/HA%20INTRO.htm#CONT
[27] American Academy of Dermatology. “How Fast Does Hair Grow? Tips for Growth,” Healthline, 2018 [online]. Available: https://www.healthline.com/health/beauty-skin-care/grow-hair-faster
[28] L.Tirado-Lee. “The science of curls,” Helix [online]. Available: https://helix.northwestern.edu/blog/2014/05/science-curls
[29] Paton, C., Hellstrom, J., Paul, B.,Woodhead, J. and Hergt, J., 2011. Iolite: Freeware for the visualisation and processing of mass spectrometric data. Journal of Analytical Atomic Spectrometry. doi:10.1039/c1ja10172b.
[30] “What is DNA?,” Genetics Home Reference, 2018 [online]. Available: https://ghr.nlm.nih.gov/primer/basics/dna.
[31] “About us,” 23andme [online]. Available: https://mediacenter.23andme.com/about-us/
[32] “Getting started with the haplogroup reports,” 23andme [online]. Available: https://customercare.23andme.com/hc/en-us/articles/235201447-Getting-Started-with-the-Haplogroup-Reports