Projects:2019s1-141 CSI Adelaide: Who killed the Somerton Man?
Contents
Project Team
Project Members
1. Azizul Hakim Luqman Ul Hakim Ng 2. Zihe Wang
Project Supervisors
1. Prof. Derek Abbott 2. Dr. Andrew Allison
Abstract
This project investigates the DNA of a man who was found dead at the Somerton Beach, South Australia, in 1948. The cause of his death remains unknown until now and police could not find any identity of that man during his death. This project consists of several tasks that contributes to solving the mystery of the Somerton Man's identity. The DNA of the deceased man which was extracted from his hair sample from the South Australian Police Historical Society was given to this group. By using the DNA sample, some analyses were done. The first task counts the amount of SNP available in the Somerton Man's DNA file. The amount of SNP available in his DNA file is insufficient for analysis on GEDmatch, a website used to analyse DNA. Using a certain programming algorithm, a number of SNPs are added to the each chromosome in the Somerton Man's DNA file to be able to investigate his DNA using GEDmatch. The second task determines the ethnicity of the Somerton Man. This process can be done without modifying the Somerton Man's DNA file. In other words, from his original file, his ethnicity can be determined using a tool provided in GEDmatch.
Introduction
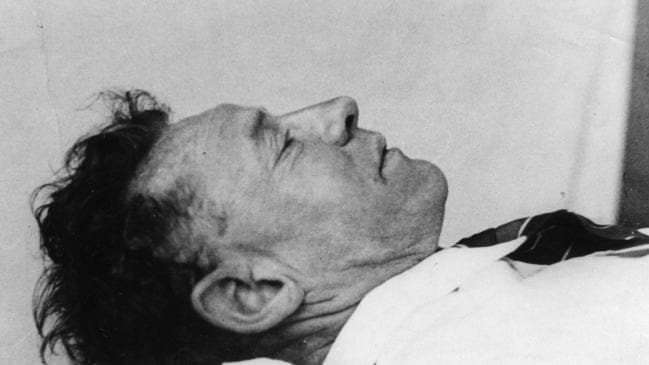
In 1948, a man was found mysteriously dead on Somerton Beach, Adelaide, South Australia. His identity remains unknown until this day, and the case has been classified as one of Australia’s biggest unsolved mysteries. There was no ID or anything on him that shows a clue on who he actually was.
This project analyses Somerton Man’s DNA file extracted from his hair which has been corrupted. The project aims to investigate the Somerton Man’s DNA with other sample DNA files via computer techniques and biological engineering methods.
Background
DNA
DNA is the hereditary material which stores the genetic information in humans [2]. There are two types of DNA in human beings, one is known as nuclear DNA which is located in cell nucleus and another type is mitochondrial DNA which is located in the mitochondria. This project only focuses on the analysis of nuclear DNA. DNA stores genetic information as a sequence built up with four types of nitrogen bases which are adenine (A), guanine (G), cytosine (C), and thymine (T) [2]. Also, a sugar molecule and a phosphate molecule are attached to each nitrogen base to form a molecule called nucleotide. The bases would pair up (A with T and C with G) and multiple nucleotides are placed in two strands to form a double helix which looks like a spiral [2]. In general, a DNA is a genetic sequence formed by multiple base pairs. The genetic instructions of building and maintaining an organism are obtained from the order of these base pairs [2]. There are about 3 billion bases in human DNA, in which more than 99% of the bases are common in all human beings, and the physiological differences among people depends on these 1% DNA.
Chromosome
Chromosome is an integrated package of DNA molecules. It has thread-like structure, and DNA molecules are coiled up around hi stones proteins to form the structure [3]. There are 23 pairs of chromosomes in human body’s cell, which is 46 chromosomes in total. 22 pairs are called autosomes which are common for both males and females and the last 23rd pair is sex chromosomes which differ males and females. In this project, the DNA data analysis would only focus on autosomes [4].
SNP
Single nucleotide polymorphisms(SNPs) are most common type of genetic variation among human beings [5]. Each SNP represents a difference in a nucleotide which is a single DNA molecule [6]. For instance, a SNP may replace a nucleotide of base guanine (G) with cytosine (C). These SNPs can be found nearly once in every 1,000 nuceotides on average in a person’s DNA. Most SNPs do not effect health of owner. However, some of these variations may associated with diseases.
DNA reference file
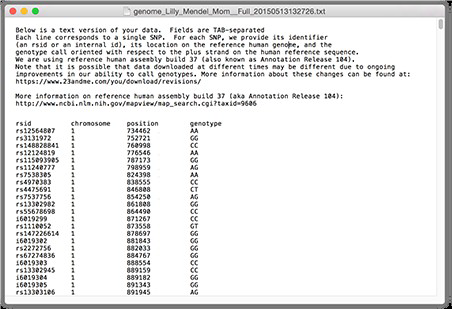
A DNA reference file stores a group of SNPs data of owner’s DNA. The format of DNA reference files using in this project is the same format of 23andMe company’s file. A screen shot of a sample file is shown below.
As shown in the figure, there are 4 columns rsid, chromosone, position and genotype in the DNA reference file. The rsid is a unique id used to identify a specific SNP [9]. The format of rsid starts with “rs” and followed by a number (eg. rs123456). These rsids are commonly used by researchers and databases. There is another special format of rsid that starts with “i” and followed by a number (eg. i123456). This “i” format is used internally by 23andMe to identify the unknown SNP and can not be used in public database. The second column chromosone identify which chromosome the SNP belongs to. Then the third column position indicates positions of SNPs in owner’s DNA sequence. Last column genotype represent the base pairs of variants(A, T, G, or C). Note that there are some cases, the genotype result for some SNPs are not able be provided and “--” would be displays in genotype column [9].
Task 1
Firstly, the Somerton man’s DNA file was examined and the available SNPs to be used for analysis were counted. There are more than 0.6 million SNPs in Somerton man’s DNA file, but only about 2%of them have determined base pairs.
Task 2
Ethnicity check via GEDmatch shows that he was North Atlantic for a proportion of more than a quarter of the chart. The second largest section shows that he was Baltic, which does not stray too much from North Atlantic region.
There is only slight change on the ethnicity regions during the degradation process. It is shown in Figure 5 that the ethnicity does not intersect with one another for two sample DNA files, thus concludes that the degradation of DNA does not affect the proportion of ethnicity. This then concludes that the Somerton Man’s origin is around North Atlantic countries and Baltic region based on Figure 4. The countries that are associated with these regions are shown in Figure 6.
Task 3
Somerton Man’s DNA was analysed with dbSNP 575 potential genetic diseases were found associated to Somerton Man’s DNA. There is no result strongly support Somerton Man's known physical appearence such as hair colour, teeth structure or eye colour. But several interesting characteristics were discovered. One of the diseases found in his DNA is Skin fragility woolly hair syndrome which indicates that Somerton Man might have woolly hair abnormality.
Conclusion
Task 1: The proportion of Somerton Man’s DNA is quite low to conduct most DNA analysis services. But there still are some techniques can be tested with it.
Task 2: The Somerton Man might be North Atlantic according to the ethnicity check on GEDmatch
Task 3: No strong evidences to confirm his physical charateristics and genetic diseases. But several interesting results were discovered.